A motif is generated from the specified IUPAC sequence fragment
by distributing the site counts equally over each possible residue. The pseudocount is added to each residue in proportion to
the background. Thus the total count for a position is the site count plus the pseudocount. For example suppose one of the
IUPAC codes entered is 'R'. The IUPAC code 'R' means that both Adenine and Guanine are possible, so with a site count of 10,
pseudocount of 1 and a uniform background; the probability of Adenine and Guanine would be:
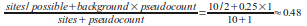
while the probility of Cytosine and Thymine would be:
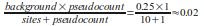
[
close]