QC statistics for lane 6
| Date |
Flowcell |
Lane |
Platform Unit |
Readgroup |
Sample |
Library |
Type |
Project |
Genome |
Centre |
|
B04DPABXX |
6 |
GA-DATA_6 |
WTCHG_11825 |
samples |
040/11 |
gDNA PE |
P100244 |
Mouse37 |
WTCHG |
| Lane |
Length |
Tiles |
Clusters PF |
% PF |
Yield (Mrd) |
Yield (Mb) |
Yield (Mb Q20) |
% Mapped |
% Coverage⊥ |
% Primer |
% Variants |
Mean cov.* |
% high cov.ℵ |
% dups |
% pair dups |
Link |
| 6.1 |
101 |
32 |
2817205 |
88.4 |
79.66 |
8045.29 |
7643.55 |
98.8 |
83.7 |
0.00 |
0.89 ± 0.00 |
3.48 |
13.68 |
6.31 |
5.44 |
back |
| 6.2 |
101 |
32 |
2817205 |
88.4 |
79.66 |
8045.29 |
7503.32 |
98.5 |
83.7 |
0.00 |
0.94 ± 0.00 |
3.47 |
13.71 |
6.30 |
5.45 |
|
⊥ Fraction of reference that is covered at least once * Mean coverage is computed over regions that are covered at least once ℵ Proportion of reads in regions with coverage in top 0.1 percentile
Lane QC statistics and plots
| Lane |
% GC |
% GCmapped |
σpos(%GC) |
insert ± MAD |
% exonic |
% exon cov'ge |
%N |
maxpos %N |
%lowQ |
%lowQend |
avgQ |
| 6.1 |
41.5 ± 8.1 |
41.5 ± 8.1 |
0.37 |
403 ± 36 |
1.3 |
93.0 |
0.0 |
0.1 |
3.8 |
17.7 |
31.8 |
| 6.2 |
40.5 ± 10.0 |
40.6 ± 9.7 |
0.44 |
403 ± 36 |
1.3 |
93.1 |
0.0 |
0.0 |
5.2 |
19.9 |
30.6 |
G+C histogram∋
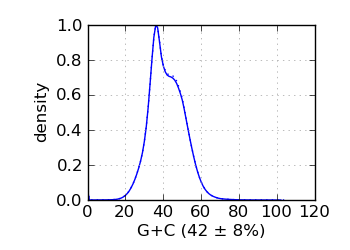 |
Insert size histogram∞
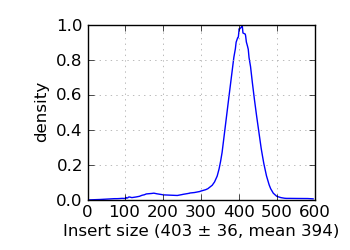 |
Mapped coverage by G+C℘
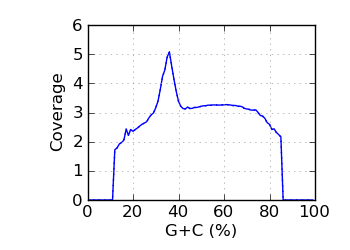 |
|
Coverage histogram
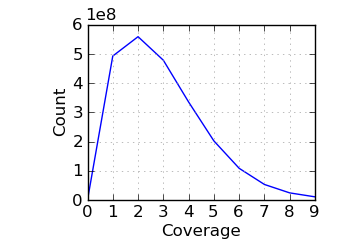 |
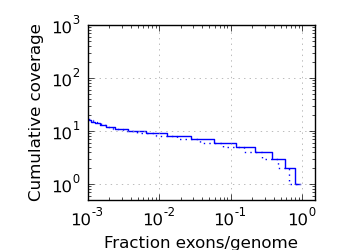
Exon/genome coverage distribution
 |
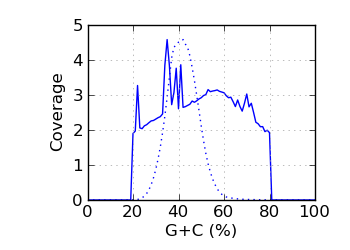
Genomic coverage by G+C∅
 |
|
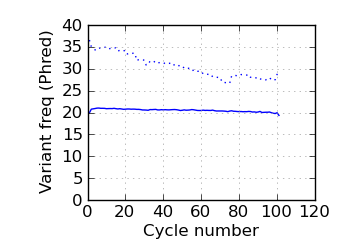
(Predicted) variants by cycle∇ (read 1)
 |
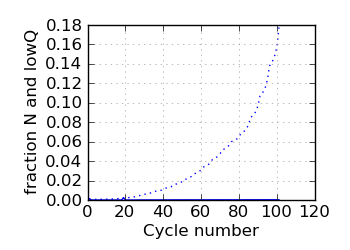
Fraction N/lowQ∋, read 1
 |
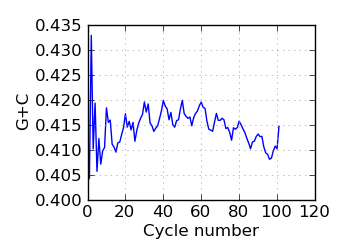
G+C by cycle (PF)∋, read 1
 |
|
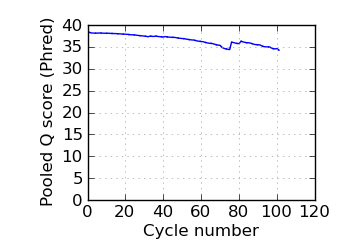
Mean Q by cycle∇, read 1
 |
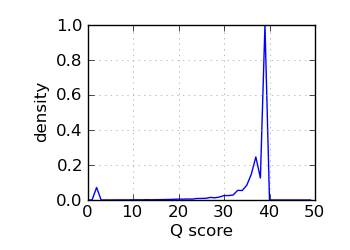
Q score histogram, read 1
 |
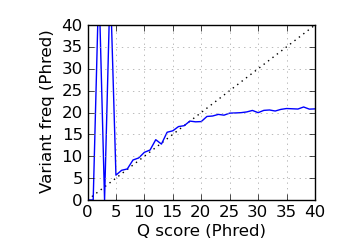
Variants by Q, read 1
 |
|
(Predicted) variants by cycle∇ (read 2)
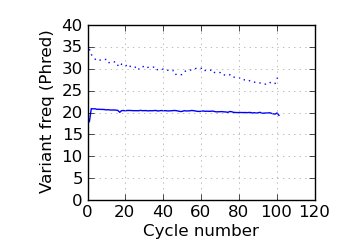 |
Fraction N/lowQ∋, read 2
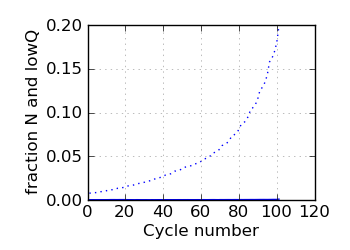 |
G+C by cycle (PF)∋, read 2
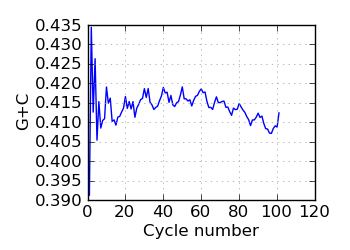 |
|
Mean Q by cycle∇, read 2
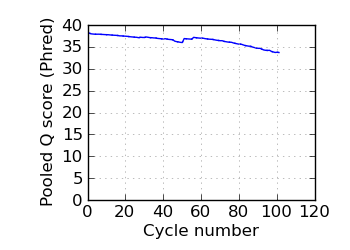 |
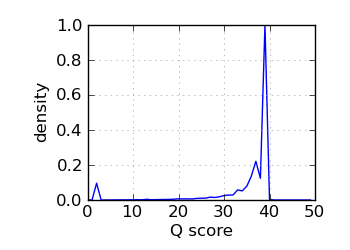
Q score histogram, read 2
 |
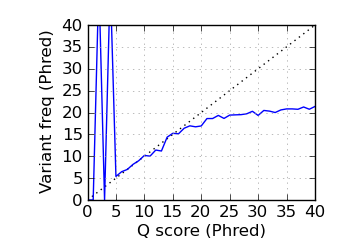
Variants by Q, read 2
 |
|
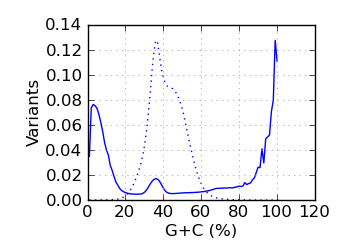
Variants by GC
 |
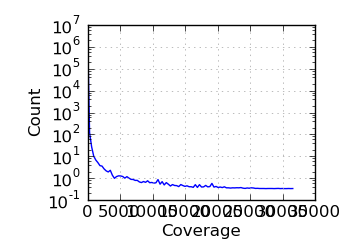
Log coverage histogram
 |
Legend
 |
∋ The Fraction N/Low Q plots, and dotted lines on the GC histogram plots, refer to all reads that have
passed chastity filters. If a reference genome was available, all others refer to mapped reads, otherwise
they too refer to chastity-filtered reads. The dotted lines in the fraction N/lowQ plot correspond to the
fraction of bases with quality score 4 or less. ∇ "Predicted variants" (dashed line) is the expected error frequency expressed as a Phred score,
and may be compared with the "Variants by cycle" graph (solid line). "Mean Q" (solid) is the numerical
mean Q score and is a measure of the average information content per read. These graphs use mapped reads only;
the dashed line in the Mean Q plot uses all (PF) reads. All four graphs are calculated on called bases with
Q Phred score above 4 only. ℘ Mapped coverage by G+C. The coverage was averaged over those genomic regions that were covered at least once.
Regions with coverage in the top 0.1 percentile were excluded; the dotted line shows results for all reads.
The G+C fraction was computed from read bases, excluding Ns and bases with quality below 4. ∅ Genomic coverage by G+C. The G+C fraction was computed from the reference genome, over the approximate fragment
Regions with coverage in the top 0.1 percentile were excluded.
The G+C histogram is shown as a dotted line (arbitrary Y scale). ∞ The insert size distribution is summarized by the median and median absolute deviation.
Multiplex QC statistics
| Lane |
Mx |
Yield Mrd |
Yield Mb Q20 |
% mapped |
% variants |
% exonic |
% exon coverage |
% pair dups |
% GC |
% GCmapped |
σpos(%GC) |
insert ± MAD |
%lowQ |
avg Q |
| 6.1 |
All |
79.66 |
7643.55 |
98.8 |
0.86 |
1.3 |
93.0 |
5.44 |
41.5 ± 8.1 |
41.5 ± 8.1 |
0.37 |
403 ± 36 |
3.8 |
31.8 |
| 6.2 |
All |
79.66 |
7503.32 |
98.5 |
0.90 |
1.3 |
93.1 |
5.45 |
40.5 ± 10.0 |
40.6 ± 9.7 |
0.44 |
403 ± 36 |
5.2 |
30.6 |
Yield (Mrd)
 |
Median insert size
 |
MAD insert size
 |
|
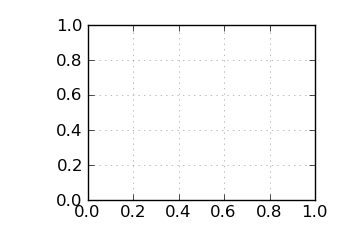
Mean G+C
 |
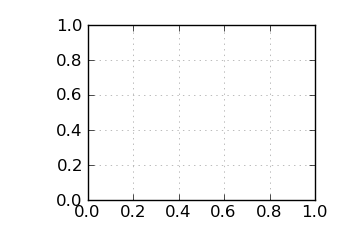
Std. dev. G+C by reads
 |
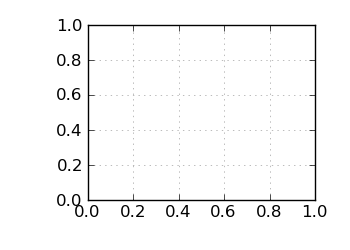
Std. dev. G+C by position
 |
Tile QC statistics and plots
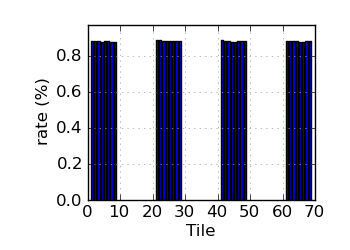
Variant rate by tile∝ (read 1)
 |
Raw/mapped yield by tile (read 1)
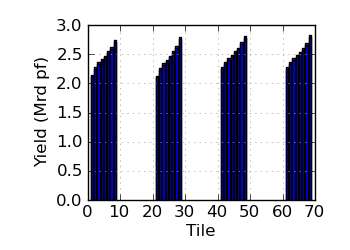 |
Fraction N/lowQ by tile (read 1)
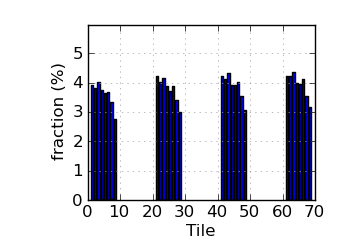 |
|
Variant rate by tile∝ (read 2)
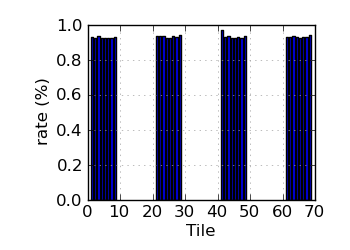 |
Raw/mapped yield by tile (read 2)
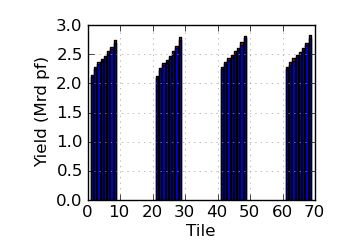 |
Fraction N/lowQ by tile (read 2)
 |
∝ HiSeq tiles are grouped in order: swathe 1 top; swathe 1 bottom; swathe 2 top; etc.
| Lane |
Tile |
Yield (pf, Mrd) |
Error rate (%) |
Mapped (%) |
Average Q |
N bases (%) |
LowQ (%) |
Last base lowQ (%) |
| 6.1 |
1 |
2.149 |
0.88 |
98.75 |
36.78 |
0.02 |
3.90 |
18.09 |
| 6.1 |
2 |
2.278 |
0.88 |
98.75 |
36.74 |
0.02 |
3.81 |
18.00 |
| 6.1 |
3 |
2.364 |
0.88 |
98.74 |
36.67 |
0.02 |
4.03 |
18.71 |
| 6.1 |
4 |
2.411 |
0.88 |
98.75 |
36.72 |
0.02 |
3.74 |
17.79 |
| 6.1 |
5 |
2.475 |
0.88 |
98.74 |
36.74 |
0.02 |
3.66 |
17.34 |
| 6.1 |
6 |
2.548 |
0.88 |
98.73 |
36.71 |
0.02 |
3.66 |
17.20 |
| 6.1 |
7 |
2.627 |
0.88 |
98.74 |
36.73 |
0.02 |
3.32 |
15.66 |
| 6.1 |
8 |
2.740 |
0.88 |
98.72 |
36.89 |
0.02 |
2.76 |
12.29 |
| 6.1 |
21 |
2.132 |
0.89 |
98.75 |
36.67 |
0.02 |
4.23 |
19.49 |
| 6.1 |
22 |
2.257 |
0.89 |
98.76 |
36.66 |
0.02 |
4.00 |
18.99 |
| 6.1 |
23 |
2.343 |
0.89 |
98.75 |
36.61 |
0.02 |
4.16 |
19.42 |
| 6.1 |
24 |
2.408 |
0.88 |
98.74 |
36.66 |
0.02 |
3.88 |
18.39 |
| 6.1 |
25 |
2.474 |
0.88 |
98.75 |
36.70 |
0.02 |
3.73 |
17.71 |
| 6.1 |
26 |
2.549 |
0.88 |
98.74 |
36.63 |
0.02 |
3.89 |
18.11 |
| 6.1 |
27 |
2.645 |
0.88 |
98.73 |
36.68 |
0.02 |
3.40 |
15.96 |
| 6.1 |
28 |
2.796 |
0.88 |
98.72 |
36.78 |
0.02 |
2.98 |
13.29 |
| 6.1 |
41 |
2.277 |
0.89 |
98.77 |
36.65 |
0.01 |
4.22 |
19.59 |
| 6.1 |
42 |
2.363 |
0.88 |
98.77 |
36.63 |
0.02 |
4.13 |
19.54 |
| 6.1 |
43 |
2.438 |
0.88 |
98.77 |
36.60 |
0.02 |
4.31 |
20.15 |
| 6.1 |
44 |
2.481 |
0.88 |
98.77 |
36.70 |
0.01 |
3.91 |
18.75 |
| 6.1 |
45 |
2.547 |
0.88 |
98.76 |
36.63 |
0.02 |
3.93 |
18.58 |
| 6.1 |
46 |
2.614 |
0.88 |
98.76 |
36.59 |
0.01 |
4.01 |
19.03 |
| 6.1 |
47 |
2.703 |
0.88 |
98.76 |
36.64 |
0.01 |
3.53 |
16.94 |
| 6.1 |
48 |
2.815 |
0.88 |
98.74 |
36.72 |
0.02 |
3.06 |
13.99 |
| 6.1 |
61 |
2.276 |
0.88 |
98.77 |
36.67 |
0.02 |
4.23 |
19.69 |
| 6.1 |
62 |
2.357 |
0.88 |
98.77 |
36.61 |
0.02 |
4.22 |
20.05 |
| 6.1 |
63 |
2.427 |
0.88 |
98.76 |
36.59 |
0.02 |
4.36 |
20.42 |
| 6.1 |
64 |
2.480 |
0.88 |
98.79 |
36.67 |
0.02 |
3.98 |
19.07 |
| 6.1 |
65 |
2.544 |
0.88 |
98.77 |
36.63 |
0.02 |
3.95 |
18.66 |
| 6.1 |
66 |
2.611 |
0.88 |
98.75 |
36.57 |
0.02 |
4.13 |
19.30 |
| 6.1 |
67 |
2.696 |
0.88 |
98.76 |
36.63 |
0.02 |
3.56 |
16.88 |
| 6.1 |
68 |
2.831 |
0.88 |
98.73 |
36.72 |
0.02 |
3.17 |
14.15 |
|
| Lane |
Tile |
Yield (pf, Mrd) |
Error rate (%) |
Mapped (%) |
Average Q |
N bases (%) |
LowQ (%) |
Last base lowQ (%) |
| 6.2 |
1 |
2.149 |
0.93 |
98.49 |
36.60 |
0.04 |
5.04 |
20.39 |
| 6.2 |
2 |
2.278 |
0.93 |
98.52 |
36.59 |
0.04 |
5.09 |
20.44 |
| 6.2 |
3 |
2.364 |
0.94 |
98.45 |
36.54 |
0.04 |
5.38 |
20.28 |
| 6.2 |
4 |
2.411 |
0.92 |
98.51 |
36.64 |
0.04 |
4.91 |
19.38 |
| 6.2 |
5 |
2.475 |
0.93 |
98.49 |
36.64 |
0.04 |
4.94 |
18.90 |
| 6.2 |
6 |
2.548 |
0.93 |
98.48 |
36.60 |
0.04 |
5.13 |
19.09 |
| 6.2 |
7 |
2.627 |
0.92 |
98.52 |
36.67 |
0.04 |
4.63 |
17.21 |
| 6.2 |
8 |
2.740 |
0.93 |
98.42 |
36.72 |
0.04 |
4.36 |
14.86 |
| 6.2 |
21 |
2.132 |
0.94 |
98.48 |
36.49 |
0.04 |
5.23 |
21.53 |
| 6.2 |
22 |
2.257 |
0.93 |
98.52 |
36.51 |
0.04 |
5.28 |
21.58 |
| 6.2 |
23 |
2.343 |
0.94 |
98.46 |
36.45 |
0.04 |
5.60 |
21.44 |
| 6.2 |
24 |
2.408 |
0.93 |
98.51 |
36.58 |
0.04 |
5.06 |
20.23 |
| 6.2 |
25 |
2.474 |
0.93 |
98.50 |
36.55 |
0.04 |
5.11 |
19.77 |
| 6.2 |
26 |
2.549 |
0.94 |
98.48 |
36.52 |
0.04 |
5.37 |
19.97 |
| 6.2 |
27 |
2.645 |
0.93 |
98.52 |
36.61 |
0.04 |
4.71 |
18.02 |
| 6.2 |
28 |
2.796 |
0.94 |
98.35 |
36.59 |
0.04 |
4.62 |
15.70 |
| 6.2 |
41 |
2.277 |
0.97 |
98.48 |
36.42 |
0.04 |
5.58 |
22.45 |
| 6.2 |
42 |
2.363 |
0.93 |
98.50 |
36.47 |
0.04 |
5.53 |
22.20 |
| 6.2 |
43 |
2.438 |
0.94 |
98.44 |
36.41 |
0.04 |
5.88 |
21.92 |
| 6.2 |
44 |
2.481 |
0.93 |
98.52 |
36.51 |
0.04 |
5.41 |
21.02 |
| 6.2 |
45 |
2.547 |
0.93 |
98.49 |
36.49 |
0.04 |
5.53 |
20.83 |
| 6.2 |
46 |
2.614 |
0.93 |
98.47 |
36.44 |
0.04 |
5.76 |
21.10 |
| 6.2 |
47 |
2.703 |
0.93 |
98.53 |
36.53 |
0.04 |
5.07 |
19.08 |
| 6.2 |
48 |
2.815 |
0.94 |
98.39 |
36.52 |
0.04 |
4.88 |
16.87 |
| 6.2 |
61 |
2.276 |
0.93 |
98.51 |
36.41 |
0.04 |
5.58 |
22.81 |
| 6.2 |
62 |
2.357 |
0.93 |
98.51 |
36.44 |
0.04 |
5.59 |
22.60 |
| 6.2 |
63 |
2.427 |
0.94 |
98.44 |
36.36 |
0.04 |
5.96 |
22.46 |
| 6.2 |
64 |
2.480 |
0.93 |
98.53 |
36.48 |
0.04 |
5.49 |
21.56 |
| 6.2 |
65 |
2.544 |
0.93 |
98.50 |
36.47 |
0.04 |
5.59 |
20.99 |
| 6.2 |
66 |
2.611 |
0.93 |
98.47 |
36.44 |
0.04 |
5.75 |
21.08 |
| 6.2 |
67 |
2.696 |
0.93 |
98.53 |
36.54 |
0.04 |
5.03 |
19.12 |
| 6.2 |
68 |
2.831 |
0.94 |
98.36 |
36.53 |
0.05 |
4.82 |
16.42 |
|
QC version: 2.1





















































